Background and Objective
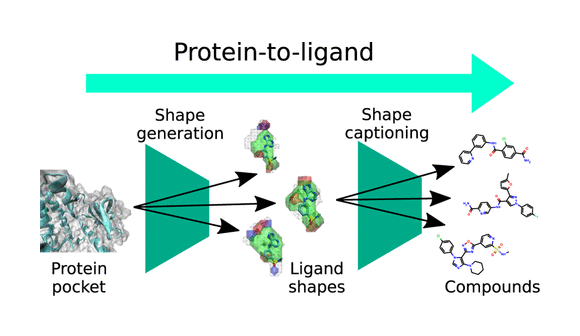
The exploration of chemical space for effective bioactive molecules poses significant challenges, particularly due to its vastness and the limitations of conventional structure-based virtual screening techniques. This case study presents LiGANN, an innovative approach using a generative adversarial network (GAN) to create diverse three-dimensional ligand shapes complementary to protein pockets, rather than relying on predefined compound libraries.
Approach and Methodology
LiGANN employs deep learning models, particularly a GAN, to generate ligand shapes based on protein structure data. The key steps involve:
- Training on Structures: Utilizing protein structures from the DUD-E dataset with known binders as generation targets, where known binders docked to each protein were used in the training.
- Featurization: Implementing a voxelization process for both ligands and proteins, with distinct channels for different atom types and properties.
- Network Architecture: Adapting BicycleGAN’s framework for three-dimensional input, using U-Net for the generator, patchGAN sequential convolutional neural networks for discriminators, and an encoder for latent code generation.
- Shape Generation: Producing diverse ligand shapes complementary to protein pockets by varying latent code values, generating more distinct and pharmacologically relevant shapes compared to previous methods like LigVoxel.
- Decoding Shapes into SMILES: Using a shape-captioning network to convert the generated shapes into SMILES strings, representing valid molecular structures.
Results
- Diversity and Relevance: LiGANN successfully generated a diverse range of ligand shapes, capable of accommodating various pharmacophoric models and compound scaffolds.
- Efficient Decoding: The method effectively decoded generated shapes into SMILES, resulting in a high success rate of grammatically correct and valid molecular structures.
- QSAR Evaluation: LiGANN compounds showed enrichment in QSAR model predictions, indicating potential as effective binders, with a notable enrichment in the top 1% of compounds.
Case Analysis: Drug Targets
LiGANN was applied to specific drug targets like delta opioid 7TM receptor, Serine/threonine-protein kinase CHK1, and Serine/threonine-protein kinase TNNI3K. The generated compounds were evaluated for their fit in the protein binding pockets and compared against known binders. The analysis demonstrated LiGANN's capability to produce compounds with similar structural features to known binders, highlighting its potential in identifying novel compounds with significant bioactivity.
Conclusion
LiGANN represents a significant advancement in the field of structure-based de novo drug design. Its ability to generate diverse and pharmacologically relevant ligand shapes, combined with efficient decoding into SMILES, offers a promising approach for exploring larger regions of chemical space. This technology not only enhances the drug discovery process but also opens up new avenues for creating novel compounds that could lead to effective treatments for various diseases.


